🟠 Brusselator 1d with periodic BC using FourierFlows.jl
The example was done in collaboration with Navid C. Constantinou.
We look at the Brusselator in 1d, see [Tzou]. The equations are
\[\begin{aligned} \frac { \partial u } { \partial t } & = D \frac { \partial ^ { 2 } u } { \partial z ^ { 2 } } + u ^ { 2 } v - ( B + 1 ) u + E \\ \frac { \partial v } { \partial t } & = \frac { \partial ^ { 2 } v} { \partial z ^ { 2 } } + B u - u ^ { 2 } v \end{aligned}\tag{E}\]
with periodic boundary conditions. These equations have been introduced to reproduce an oscillating chemical reaction.
We focus on computing a snaking branch of periodic orbits using spectral methods implemented in Brusselator.jl:
using Revise, BifurcationKit
using Brusselator, Plots, Parameters, ForwardDiff, LinearAlgebra, Setfield, DiffEqBase
using FFTW: irfft
const BK = BifurcationKit
dev = CPU() # Device (CPU/GPU)
nx = 512 # grid resolution
stepper = "FilteredRK4" # timestepper
dt = 0.01 # timestep
nsteps = 9000 # total number of time-steps
nsubs = 20 # number of time-steps for intermediate logging/plotting (nsteps must be multiple of nsubs)
# parameters for the model used by Tzou et al. (2013)
E = 1.4
L = 137.37 # Domain length
ε = 0.1
μ = 25
ρ = 0.178
D_c = ((sqrt(1 + E^2) - 1) / E)^2
B_H = (1 + E * sqrt(D_c))^2
B = B_H + ε^2 * μ
D = D_c + ε^2 * ρ
kc = sqrt(E / sqrt(D))
# building the model
grid = OneDGrid(dev, nx, L)
params = Brusselator.Params(B, D, E)
vars = Brusselator.Vars(dev, grid)
equation = Brusselator.Equation(dev, params, grid)
prob = FourierFlows.Problem(equation, stepper, dt, grid, vars, params, dev)
get_u(prob) = irfft(prob.sol[:, 1], prob.grid.nx)
get_v(prob) = irfft(prob.sol[:, 2], prob.grid.nx)
u_solution = Diagnostic(get_u, prob; nsteps=nsteps+1)
diags = [u_solution]We now integrate the model to find a periodic orbit:
l = 28
θ = @. (grid.x > -l/2) & (grid.x < l/2)
au, av = -(E^2 + kc^2) / B, -1
cu, cv = -E * (E + im) / B, 1
# initial condition
u0 = @. E + ε * real( au * exp(im * kc * grid.x) * θ + cu * (1 - θ) )
v0 = @. B/E + ε * real( av * exp(im * kc * grid.x) * θ + cv * (1 - θ) )
set_uv!(prob, u0, v0)
plot_output(prob)
# move forward in time to capture the periodic orbit
for j=0:Int(nsteps/nsubs)
updatevars!(prob)
stepforward!(prob, diags, nsubs)
end
# estimate of the periodic orbit, will be used as initial condition for a Krylov-Newton
initpo = copy(vcat(vcat(prob.vars.u, prob.vars.v), 4.9))
using RecursiveArrayTools
t = u_solution.t[1:10:nsteps]
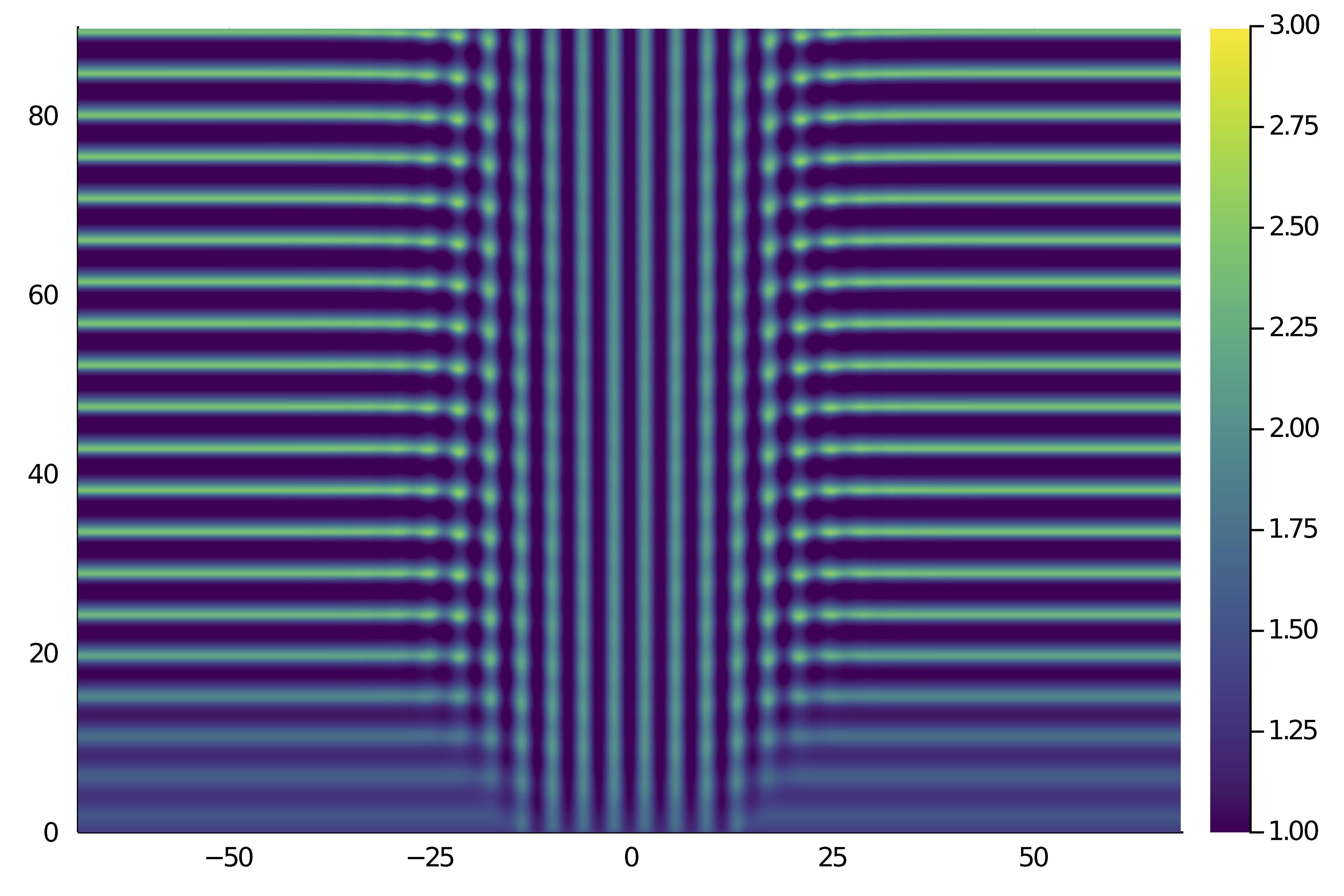
U_xt = reduce(hcat, [ u_solution.data[j] for j=1:10:nsteps ])
heatmap(grid.x, t, U_xt', c = :viridis, clims = (1, 3))which gives
Building the Shooting problem
We compute the periodic solution of (E) with a shooting algorithm. We thus define a function to compute the flow and its differential.
# update the states
function _update!(out, pb::FourierFlows.Problem, N)
out[1:N] .= pb.vars.u
out[N+1:end] .= pb.vars.v
out
end
# update the parameters in pb
function _setD!(D, pb::FourierFlows.Problem)
pb.eqn.L[:, 1] .*= D / prob.params.D
pb.params.D = D
end
# compute the flow from x up to time t
function ϕ(x, p, t)
@unpack pb, D, N = p
_setD!(D, pb)
# set initial condition
@views set_uv!(pb, x[1:N], x[N+1:2N])
pb.clock.t=0.; pb.clock.step=0
# determine number of time steps
dt = pb.clock.dt
nsteps = div(t, dt) |> Int
# compute flow
stepforward!(pb, nsteps)
rest = t - nsteps * dt
dt = pb.clock.dt
pb.clock.dt = rest
stepforward!(pb, 1)
pb.clock.dt = dt
updatevars!(pb)
out = similar(x)
_update!(out, pb, N)
return (t=t, u=out)
end
# differential of the flow by FD
function dϕ(x, p, dx, t; δ = 1e-8)
phi = ϕ(x, p, t).u
dphi = (ϕ(x .+ δ .* dx, p, t).u .- phi) ./ δ
return (t=t, u=phi, du=dphi)
endWe also need the vector field
function vf(x, p)
@unpack pb, D, N = p
# set parameter in prob
pb.eqn.L[:, 1] .*= D / pb.params.D
pb.params.D = D
u = @view x[1:N]
v = @view x[N+1:end]
# set initial condition
set_uv!(pb, u, v)
rhs = Brusselator.get_righthandside(pb)
# rhs is in Fourier space, put back to real space
out = similar(x)
ldiv!((@view out[1:N]), pb.grid.rfftplan, rhs[:, 1])
ldiv!((@view out[N+1:end]), pb.grid.rfftplan, rhs[:, 2])
return out
endWe then specify the shooting problem
# parameters to be passed to ϕ
par_bru = (pb = prob, N = nx, nsubs = nsubs, D = D)
# example of vector passed to ϕ
x0 = vcat(u0,v0)
# here we define the problem which encodes the standard shooting
flow = Flow(vf, ϕ, dϕ)
# the first section is centered around a stationary state
_center = vcat(E*ones(nx), B/E*ones(nx))
# section for the flow
sectionBru = BK.SectionSS(vf(initpo[1:end-1], par_bru), _center)
probSh = ShootingProblem(M = 1, flow = flow, ds = diff(LinRange(0, 1, 1 + 1)), section = sectionBru, par = par_bru, lens = (@lens _.D), jacobian = :FiniteDifferences)Finding a periodic orbit
# linear solver for the Shooting problem
ls = GMRESIterativeSolvers(N = 2nx+1, reltol = 1e-4)
# parameters for Krylov-Newton
optn = NewtonPar(tol = 1e-9, verbose = true,
# linear solver
linsolver = ls,
# eigen solver
eigsolver = EigKrylovKit(dim = 30, x₀ = rand(2nx), verbose = 1 )
)
# Newton-Krylov method to check convergence and tune parameters
solp = @time newton(probSh, initpo, optn,
normN = x->norm(x,Inf))and you should see (the guess was not that good)
┌─────────────────────────────────────────────────────┐
│ Newton step residual linear iterations │
├─────────────┬──────────────────────┬────────────────┤
│ 0 │ 1.6216e+03 │ 0 │
│ 1 │ 4.4376e+01 │ 6 │
│ 2 │ 9.0109e-01 │ 15 │
│ 3 │ 4.9044e-02 │ 32 │
│ 4 │ 4.1139e-03 │ 27 │
│ 5 │ 6.0704e-03 │ 33 │
│ 6 │ 2.4721e-04 │ 33 │
│ 7 │ 3.7889e-05 │ 34 │
│ 8 │ 6.6836e-07 │ 31 │
│ 9 │ 2.0295e-09 │ 39 │
│ 10 │ 2.1685e-12 │ 41 │
└─────────────┴──────────────────────┴────────────────┘
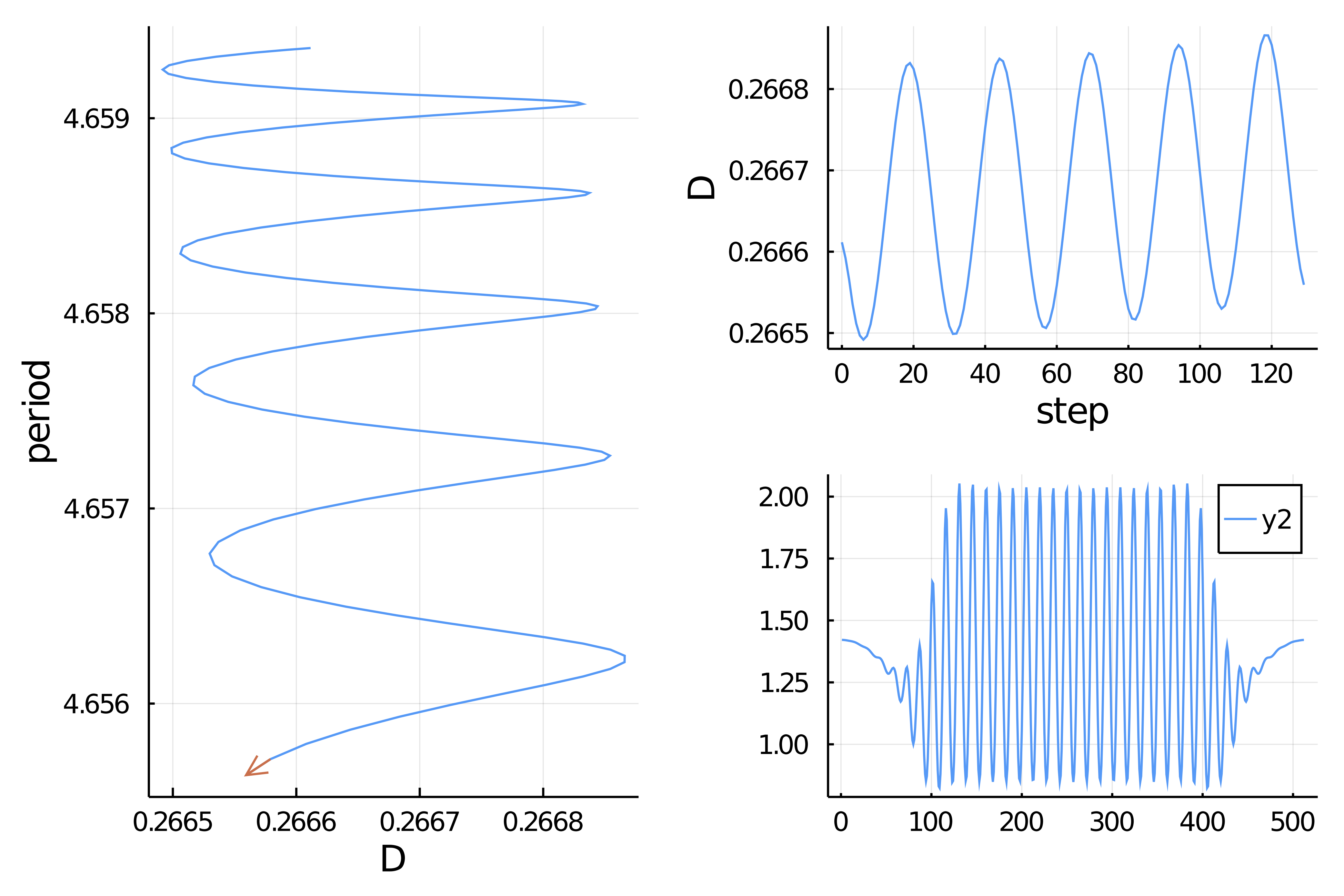
83.615047 seconds (284.31 M allocations: 24.250 GiB, 6.28% gc time)Computation of the snaking branch
# you can detect bifurcations with the option detect_bifurcation = 3
optc = ContinuationPar(newton_options = optn, ds = -1e-3, dsmin = 1e-7, dsmax = 2e-3, p_max = 0.295, plot_every_step = 2, max_steps = 1000, detect_bifurcation = 0)
bd = continuation(probSh,
solp, PALC(bls = MatrixFreeBLS(@set ls.N = 2nx+2)), optc;
plot = true,
verbosity = 3,
plot_solution = (x,p ; kw...) -> plot!(x[1:nx];kw...),
normC = x->norm(x,Inf))which leads to
References
- Tzou
Tzou, J. C., Y.-P. Ma, A. Bayliss, B. J. Matkowsky, and V. A. Volpert. Homoclinic Snaking near a Codimension-Two Turing-Hopf Bifurcation Point in the Brusselator Model.” Physical Review E 87, no. 2 (February 14, 2013): 022908. https://doi.org/10.1103/PhysRevE.87.022908.